Publications
2024

Building Blocks of Understanding: Constructing a Reverse Genetics Platform for studying determinants of SARS-CoV-2 replication
Cold Spring Harbor Laboratory
·
06 Feb 2024
·
doi:10.1101/2024.02.05.578560

SHIFTR enables the unbiased identification of proteins bound to specific RNA regions in live cells
Nucleic Acids Research
·
28 Jan 2024
·
doi:10.1093/nar/gkae038

Nucleolar detention of NONO shields DNA double-strand breaks from aberrant transcripts
Nucleic Acids Research
·
15 Jan 2024
·
doi:10.1093/nar/gkae022
2023

SND1 binds SARS-CoV-2 negative-sense RNA and promotes viral RNA synthesis through NSP9
Cell
·
01 Oct 2023
·
doi:10.1016/j.cell.2023.09.002

Lab-scale siRNA and mRNA LNP manufacturing by various microfluidic mixing techniques – an evaluation of particle properties and efficiency
OpenNano
·
01 Jul 2023
·
doi:10.1016/j.onano.2023.100161

Codon affinity in mitochondrial DNA shapes evolutionary and somatic fitness
Cold Spring Harbor Laboratory
·
23 Apr 2023
·
doi:10.1101/2023.04.23.537997
2022

Congenital anemia reveals distinct targeting mechanisms for master transcription factor GATA1
Blood
·
21 Apr 2022
·
doi:10.1182/blood.2021013753
Protective immune trajectories in early viral containment of non-pneumonic SARS-CoV-2 infection
Nature Communications
·
23 Feb 2022
·
doi:10.1038/s41467-022-28508-0
2021

The Zinc Finger Antiviral Protein ZAP Restricts Human Cytomegalovirus and Selectively Binds and Destabilizes Viral
UL4
/
UL5
Transcripts
mBio
·
29 Jun 2021
·
doi:10.1128/mbio.02683-20

Atlas der SARS-CoV-2-RNA-Protein-Interaktionen in infizierten Zellen
BIOspektrum
·
01 Jun 2021
·
doi:10.1007/s12268-021-1587-3
2020
The SARS-CoV-2 RNA–protein interactome in infected human cells
Nature Microbiology
·
21 Dec 2020
·
doi:10.1038/s41564-020-00846-z
The lncRNA lincNMR regulates nucleotide metabolism via a YBX1 - RRM2 axis in cancer
Nature Communications
·
25 Jun 2020
·
doi:10.1038/s41467-020-17007-9
Control of human hemoglobin switching by LIN28B-mediated regulation of BCL11A translation
Nature Genetics
·
20 Jan 2020
·
doi:10.1038/s41588-019-0568-7
2019

Context-specific regulation of cell survival by a miRNA-controlled BIM rheostat
Genes & Development
·
07 Nov 2019
·
doi:10.1101/gad.330134.119
2018
New insights into the cellular temporal response to proteostatic stress
eLife
·
12 Oct 2018
·
doi:10.7554/eLife.39054
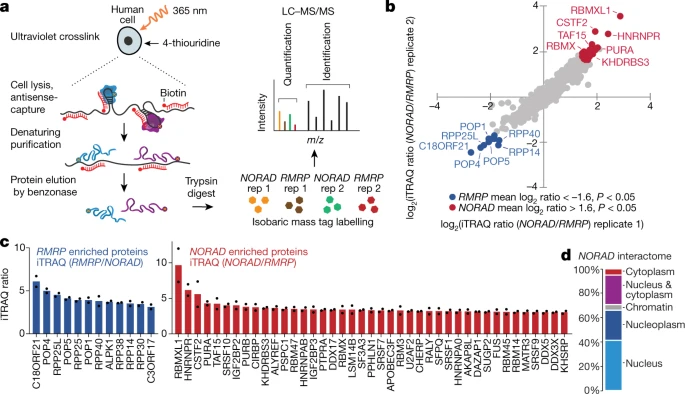
The NORAD lncRNA assembles a topoisomerase complex critical for genome stability
Nature
·
27 Aug 2018
·
doi:10.1038/s41586-018-0453-z

Nuclear lnc
RNA
stabilization in the host response to bacterial infection
The EMBO Journal
·
22 Jun 2018
·
doi:10.15252/embj.201899875

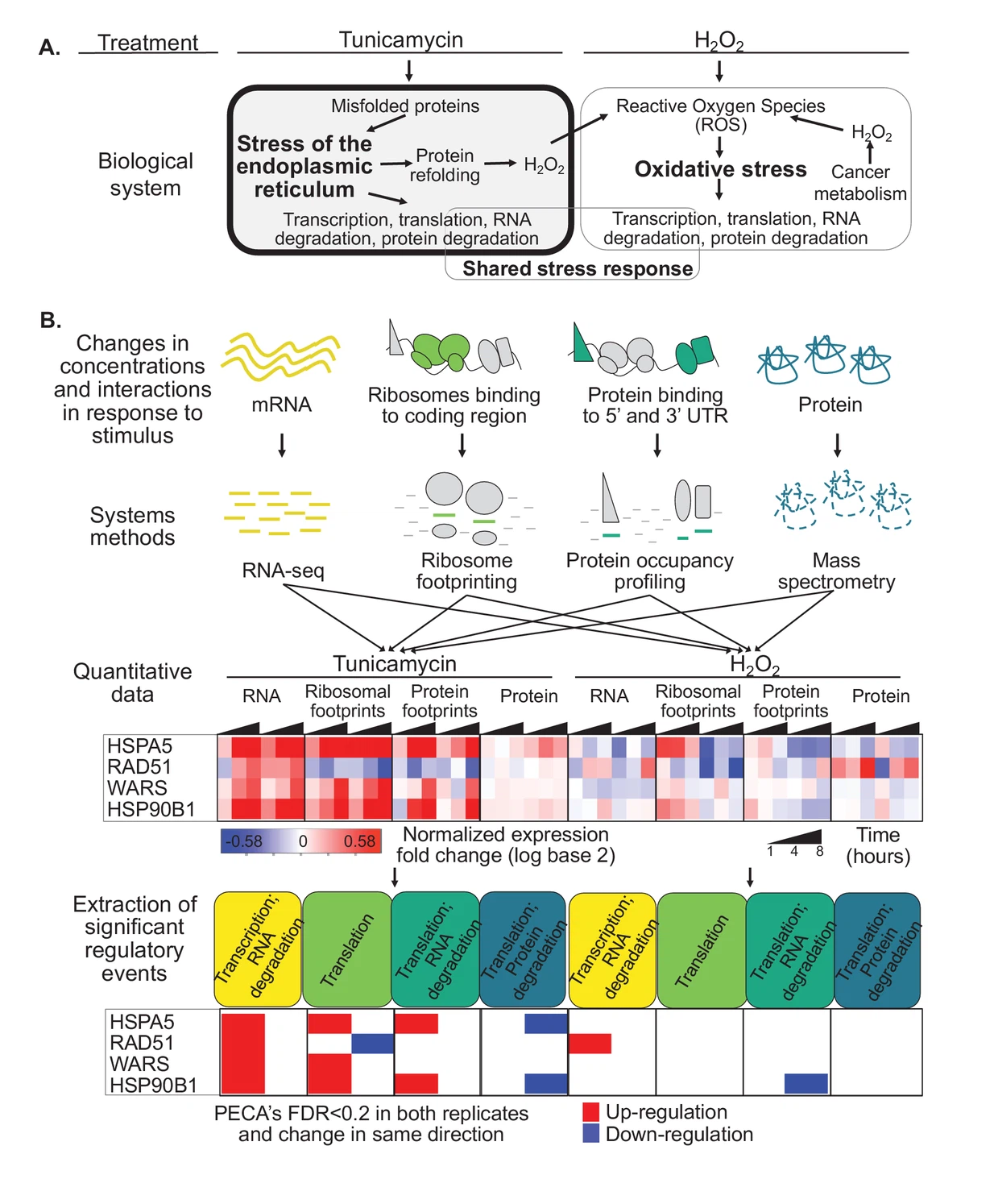
Quantifying multi-layered expression regulation in response to stress of the endoplasmic reticulum
Cold Spring Harbor Laboratory
·
25 Apr 2018
·
doi:10.1101/308379

Ribosome Levels Selectively Regulate Translation and Lineage Commitment in Human Hematopoiesis
Cell
·
01 Mar 2018
·
doi:10.1016/j.cell.2018.02.036
2017

Developmentally‐faithful and effective human erythropoiesis in immunodeficient and Kit mutant mice
American Journal of Hematology
·
19 Jul 2017
·
doi:10.1002/ajh.24805
2016

Systematic mapping of functional enhancer–promoter connections with CRISPR interference
Science
·
11 Nov 2016
·
doi:10.1126/science.aag2445
2015
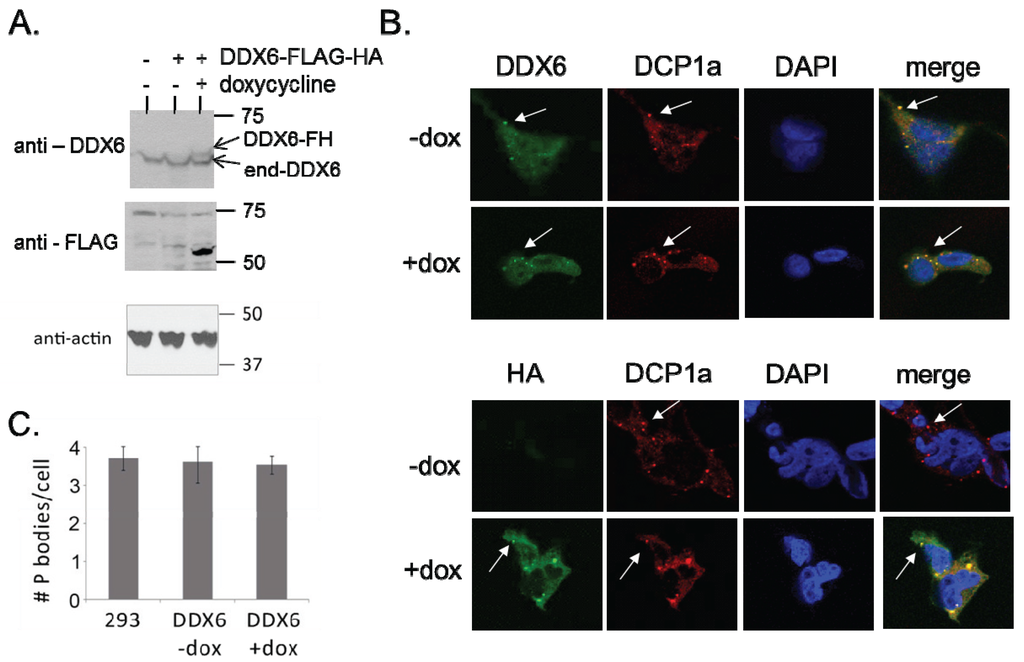
Comprehensive Protein Interactome Analysis of a Key RNA Helicase: Detection of Novel Stress Granule Proteins
Biomolecules
·
15 Jul 2015
·
doi:10.3390/biom5031441

High-Resolution Profiling of Protein-RNA Interactions
Springer Theses
·
01 Jan 2015
·
doi:10.1007/978-3-319-16253-9
2014

MOV10 Is a 5′ to 3′ RNA Helicase Contributing to UPF1 mRNA Target Degradation by Translocation along 3′ UTRs
Molecular Cell
·
01 May 2014
·
doi:10.1016/j.molcel.2014.03.017

High-resolution profiling of protein occupancy on polyadenylated RNA transcripts
Methods
·
01 Feb 2014
·
doi:10.1016/j.ymeth.2013.09.017
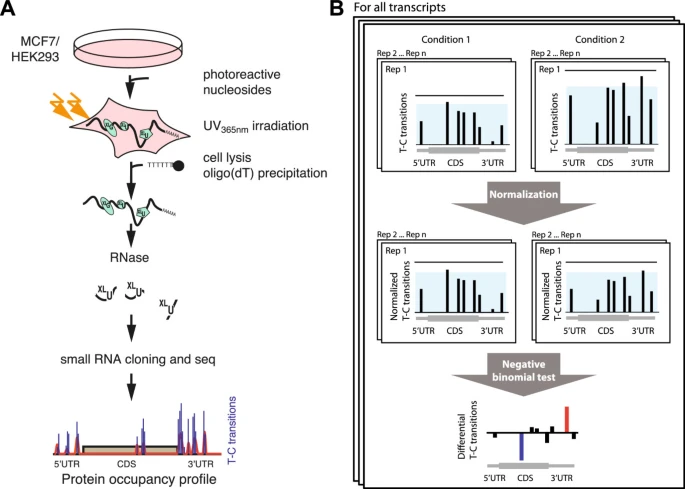
Differential protein occupancy profiling of the mRNA transcriptome
Genome Biology
·
01 Jan 2014
·
doi:10.1186/gb-2014-15-1-r15
2013

Identification of LIN28B-bound mRNAs reveals features of target recognition and regulation
RNA Biology
·
01 Jul 2013
·
doi:10.4161/rna.25194
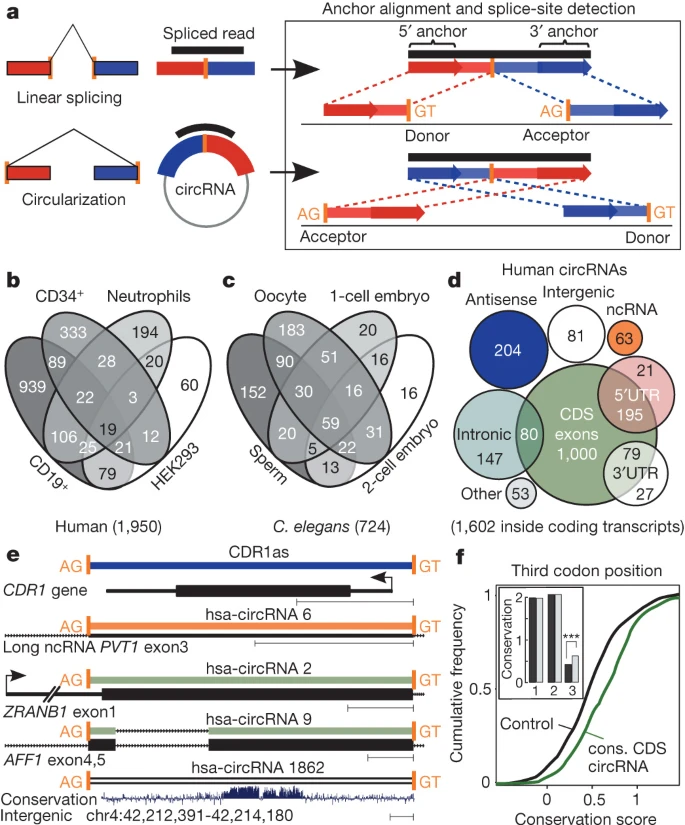
Circular RNAs are a large class of animal RNAs with regulatory potency
Nature
·
27 Feb 2013
·
doi:10.1038/nature11928
2012
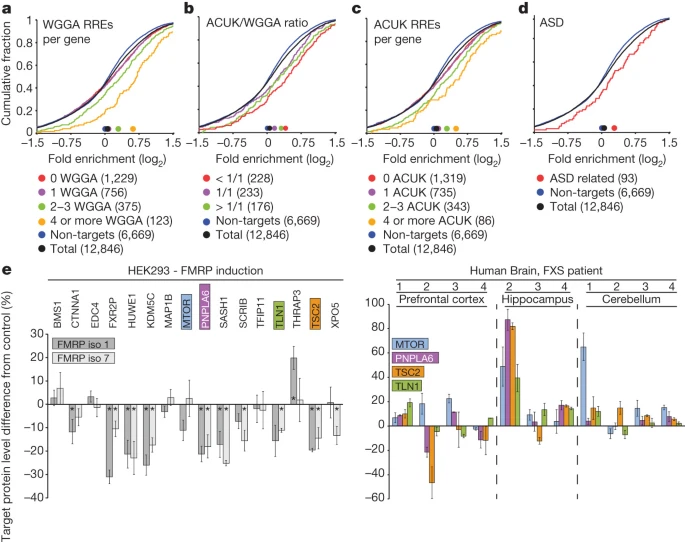
FMRP targets distinct mRNA sequence elements to regulate protein expression
Nature
·
01 Dec 2012
·
doi:10.1038/nature11737

The mRNA-Bound Proteome and Its Global Occupancy Profile on Protein-Coding Transcripts
Molecular Cell
·
01 Jun 2012
·
doi:10.1016/j.molcel.2012.05.021
2010

PAR-CliP - A Method to Identify Transcriptome-wide the Binding Sites of RNA Binding Proteins
Journal of Visualized Experiments
·
02 Jul 2010
·
doi:10.3791/2034

Transcriptome-wide Identification of RNA-Binding Protein and MicroRNA Target Sites by PAR-CLIP
Cell
·
01 Apr 2010
·
doi:10.1016/j.cell.2010.03.009